Structural Mass Spectrometry Platform
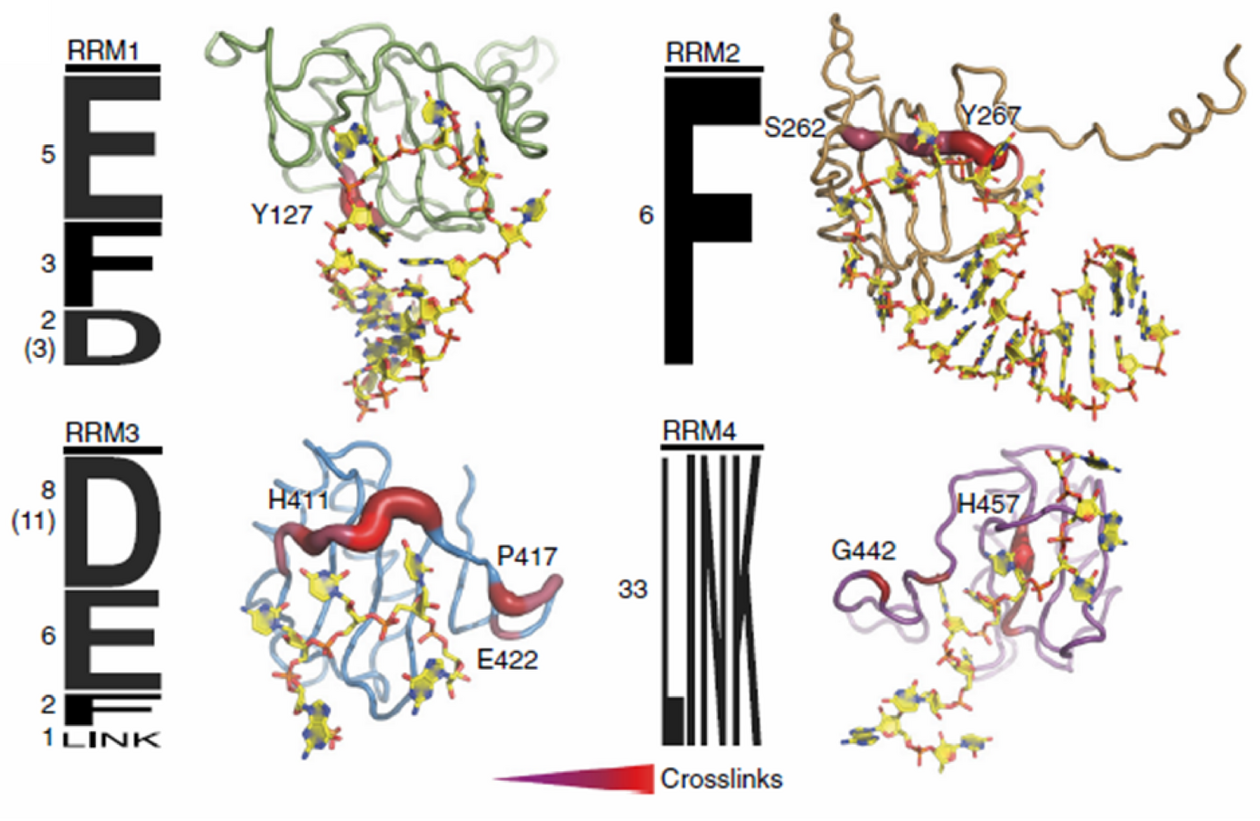
Starting from phase 2 the NCCR RNA& Disease researchers have access to a new platform for structural mass spectrometry, which will provide structural data obtained from protein-RNA and protein-protein crosslinking experiments followed by mass spectrometry analysis.
The platform is headed by Alexander Leitner, senior scientist in the group of Ruedi Aebersold at the Institute of Molecular Systems Biology at ETH Zurich. They together with the Allain lab developed the crosslinking of segmentally isotope-labeled RNA and tandem mass spectrometry (CLIR-MS/MS) method (Dorn G. et al. (2017) Nature Methods), which is now offered as a platform service. The method allows for analysis of protein-RNA interactions with single amino acid and nucleotide resolution. Given sufficient crosslinks, the method allows for structural modeling of the complexes investigated. The platform also further develops the method in close collaboration with the labs of Frederic Allain and Jonathan Hall. Researchers as well have access to chemical cross-linking of protein-protein complexes (XL-MS) for obtaining interaction information and distance restraints of multi-subunit assemblies. For example, Alexander Leitner applied XL-MS to obtain structural information on mammalian mitochondrial ribosomes in collaboration with the lab of Nenad Ban (Greber B.J. et al. (2015) Science). Researchers interested in the platform’s services can contact Alexander Leitner by email: leitner@imsb.biol.ethz.ch
By Roland Fischer
High-Throughput Crystallization Platform
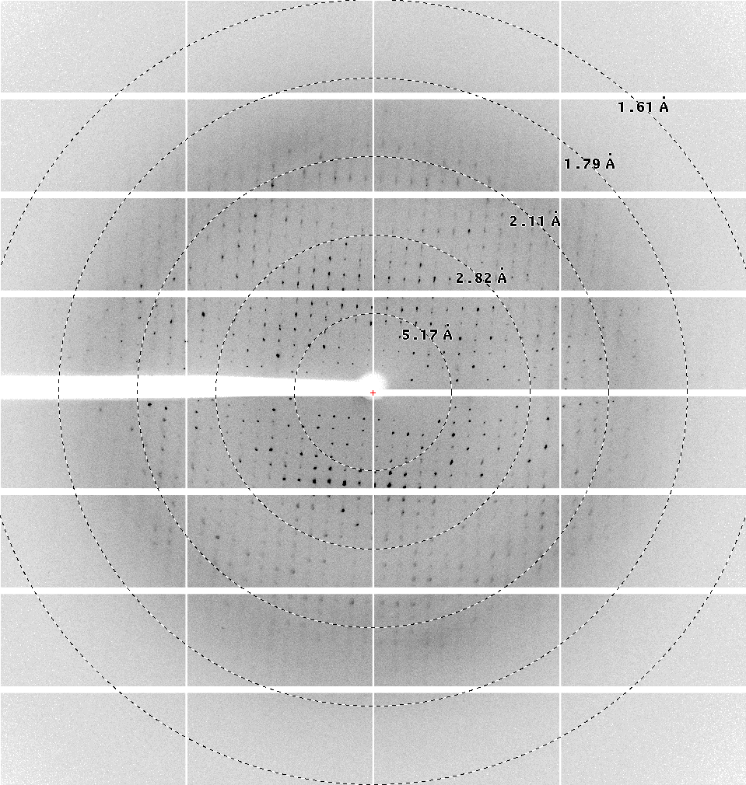
As of phase 2, the high-throughput crystallization platform of the NCCR RNA & Disease offers expanded services including remote inspection of screening experiments, the acquisition of initial synchrotron data and on a collaborative basis full structure determination.
The expanded services will be provided by the Protein Crystallization Center (PCC) and the laboratory of Martin Jinek at the Institute of Biochemistry of the University of Zurich. Users can contact the platform by sending an email to rna.xtal@bioc.uzh and registering with the PCC to get advice on sample requirements and choice of screens. Plate-based crystallization screens will be set up at the PCC by the use of pipetting robots allowing for drop volumes as low as 50 nanoliters per well and incubated in Rock Imagers with automated imaging capabilities, including UV. Images can be remotely accessed through a web interface, and the users can consult with the platform on preliminary hits obtained. Afterwards, refinement screens could be conducted, suitable crystals fished and mounted for initial synchrotron screening. Full crystal optimization and data collection with subsequent structure determination can be performed on a collaborative basis with the Jinek group. The platform’s services are free of charge except for the PCC’s screening services, which are accessible at a very favorable rate to member and associate member labs of the NCCR RNA & Disease.
Protein Crystallization Center - PCC
By Roland Fischer