Major role for minor spliceosome
Dysregulation of RNA metabolism is associated with a wide range of diseases, notably cancer and several neurological disorders. A deeper understanding of how RNA-processing is regulated could not only provide insight into the pathomechnism of diseases but also lead to the identification of targets for medical treatments.
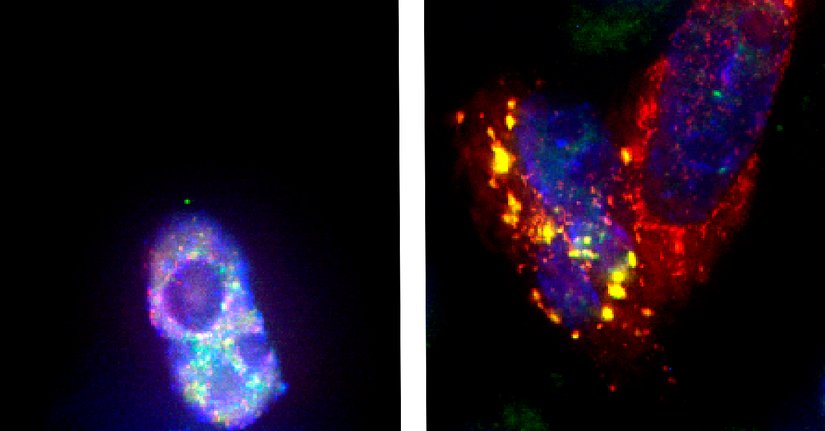
By having pinned down the mechanism of action of the RNA-binding protein Fused in Sarcoma (FUS), the groups of Marc-David Ruepp and Oliver Mühlemann now made a big step closer towards the understanding of one of the most devastating human neurological diseases, amyotrophic lateral sclerosis (ALS). Most ALS cases appear sporadic, but ALS can also be inherited (familial, fALS;~10%) due to mutations in different genes. In 2009, mutations in the FUS gene were identified as a novel cause for ALS. FUS is a ubiquitously expressed, mainly nuclear protein of the hnRNP family. Most reported ALS-causing FUS mutations are missense mutations clustered in the C-terminal nuclear localization signal, which lead to a reduced import of FUS into the nucleus and to the formation of FUS aggregates in the cytoplasm of neurons of ALS patients. Is it the loss of FUS function in the nucleus or the gain of FUS function in the cytoplasm by which mutations the FUS gene drive the pathomechanisms in ALS?
Based on more recent data showing that FUS is present in spliceosomes and that it interacts with several splicing factors, the group of Ruepp addressed the roles of FUS in influencing pre-mRNA splicing. In collaboration with researchers from the University of Milano, the Bernese characterized the FUS interactome by mass spectrometry. They found minor spliceosome components highly enriched among the FUS-interacting proteins, and that a FUS knockout affected predominantly the removal of minor introns.
The authors confirmed that FUS is necessary for regulating the splicing of minor intron-containing mRNAs, among them voltage-gated sodium channels that are required for proper muscle function and postnatal maturation of spinal motor neurons. Moreover, an ALS-associated FUS mutation that leads to cytoplasmic aggregates fails to promote minor intron splicing and traps the minor spliceosome components U11 and U12 snRNA within these aggregates.
The paper of Reber et al. shifts the attention from the current focus of the more abundant major spliceosome to the far less abundant minor spliceosome. Due to its low abundance, the minor spliceosome is much more at risk of being functionally affected by the sequestration of U11 and U12 snRNAs into cytoplasmic FUS aggregates than the major spliceosome.
The finding that FUS plays a role in splicing of minor introns suggests a possible pathomechanism for ALS and extends the spectrum of diseases, where minor spliceosome plays a major role. It even might open a new gateway to develop and design new therapeutics.
By Thomas Schnyder